|
I am a final-year PhD student at Montreal Institute for Learning Algorithms (Mila) supervised by Prof. Jian Tang. Before that, I received my B.S. in Computer Science from Peking University, advised by Prof. Ming Zhang. Email: chence.shi [at] umontreal [dot] ca |

|
|
My research is centered on the intersection of generative models, geometric deep learning, graph representation learning, and AI for science.
|
|
|
|
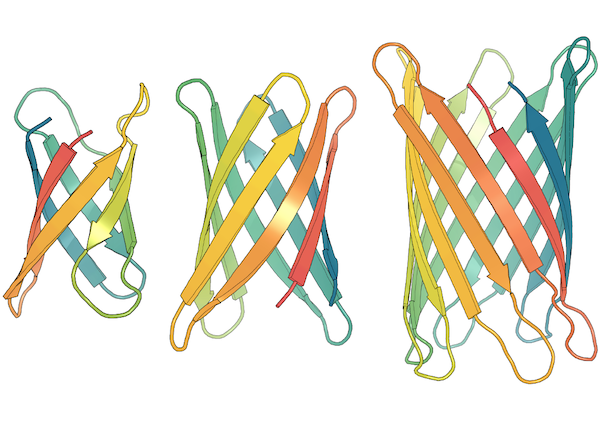
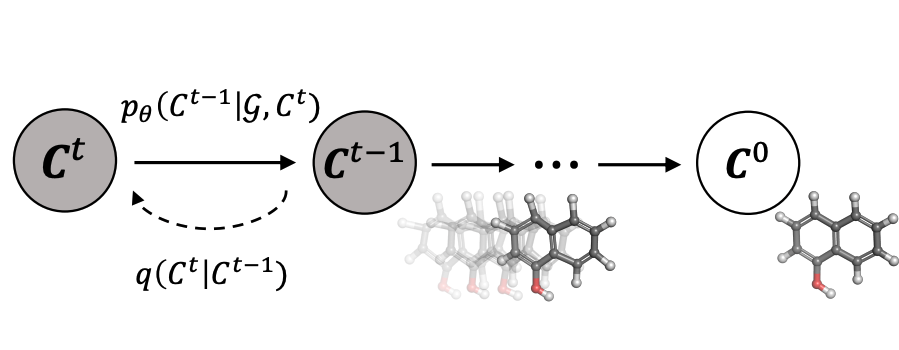
Chence Shi, Chuanrui Wang, Jiarui Lu, Bozitao Zhong, Jian Tang 11th International Conference on Learning Representations (ICLR 2023) Sequence-structure Co-Design; translation in joint sequence-structure space. [PDF] [Code] |
|
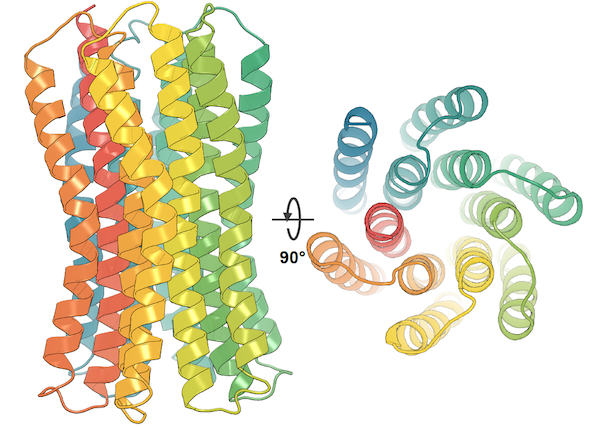
Yangtian Zhang*, Huiyu Cai*, Chence Shi, Bozitao Zhong, Jian Tang 11th International Conference on Learning Representations (ICLR 2023) End-to-end protein-ligand docking; SE(3)-equivariance. [PDF] [Code] |


|
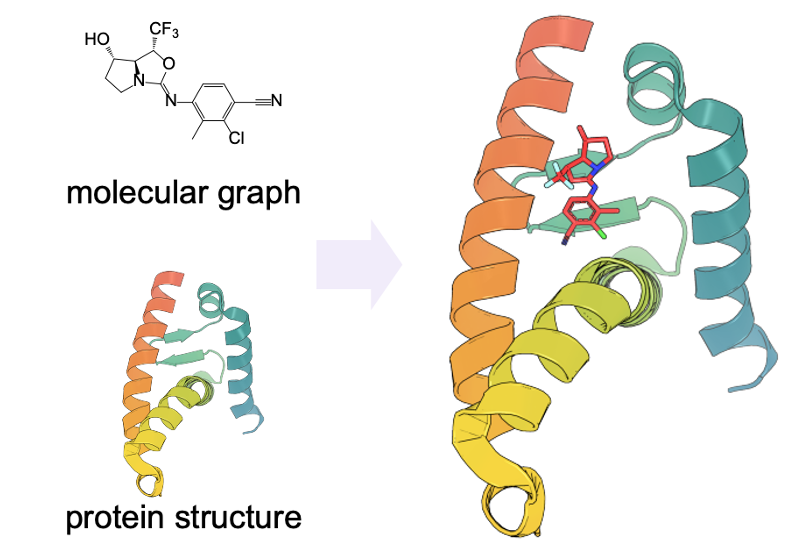
Chence Shi*, Shitong Luo*, Minkai Xu, Jian Tang 38th International Conference on Machine Learning (ICML 2021) Molecular 3D geometry generation; denoising diffusion; SE(3)-equivariance. Long talk [top 3.0%] [PDF] [Code] [Slides] |


|
Zhaocheng Zhu, Chence Shi, Zuobai Zhang, Shengchao Liu, Minghao Xu, Xinyu Yuan, Yangtian Zhang, Junkun Chen, Huiyu Cai, Jiarui Lu, Chang Ma, Runcheng Liu, Louis-Pascal Xhonneux, Meng Qu, Jian Tang Ready-to-use platfrom for fast development of drug discovery models; ML system. Deep Learning Library included in PyTorch’s ecosystem [PDF] [Homepage] [Github] [Twitter] [Google Colab] |

|
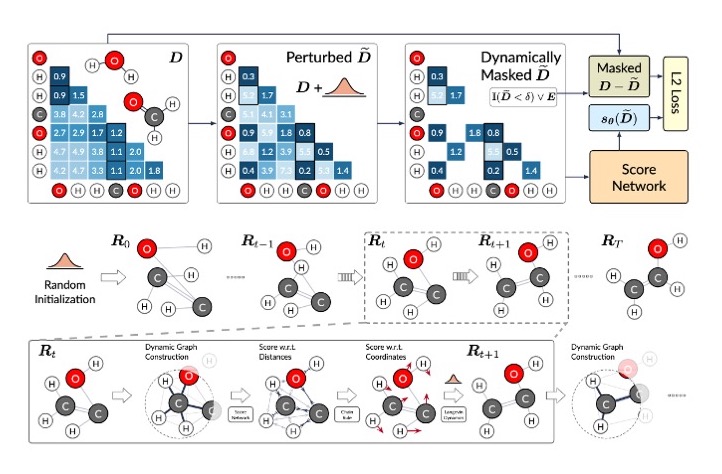
Minkai Xu, Lantao Yu, Yang Song, Chence Shi, Stefano Ermon, Jian Tang 10th International Conference on Learning Representations (ICLR 2022) Geometric probabilistic models; Markov chains; SE(3)-equivariance; denoising diffusion. Oral Presentation [54/3391] [PDF] [Code] |

|
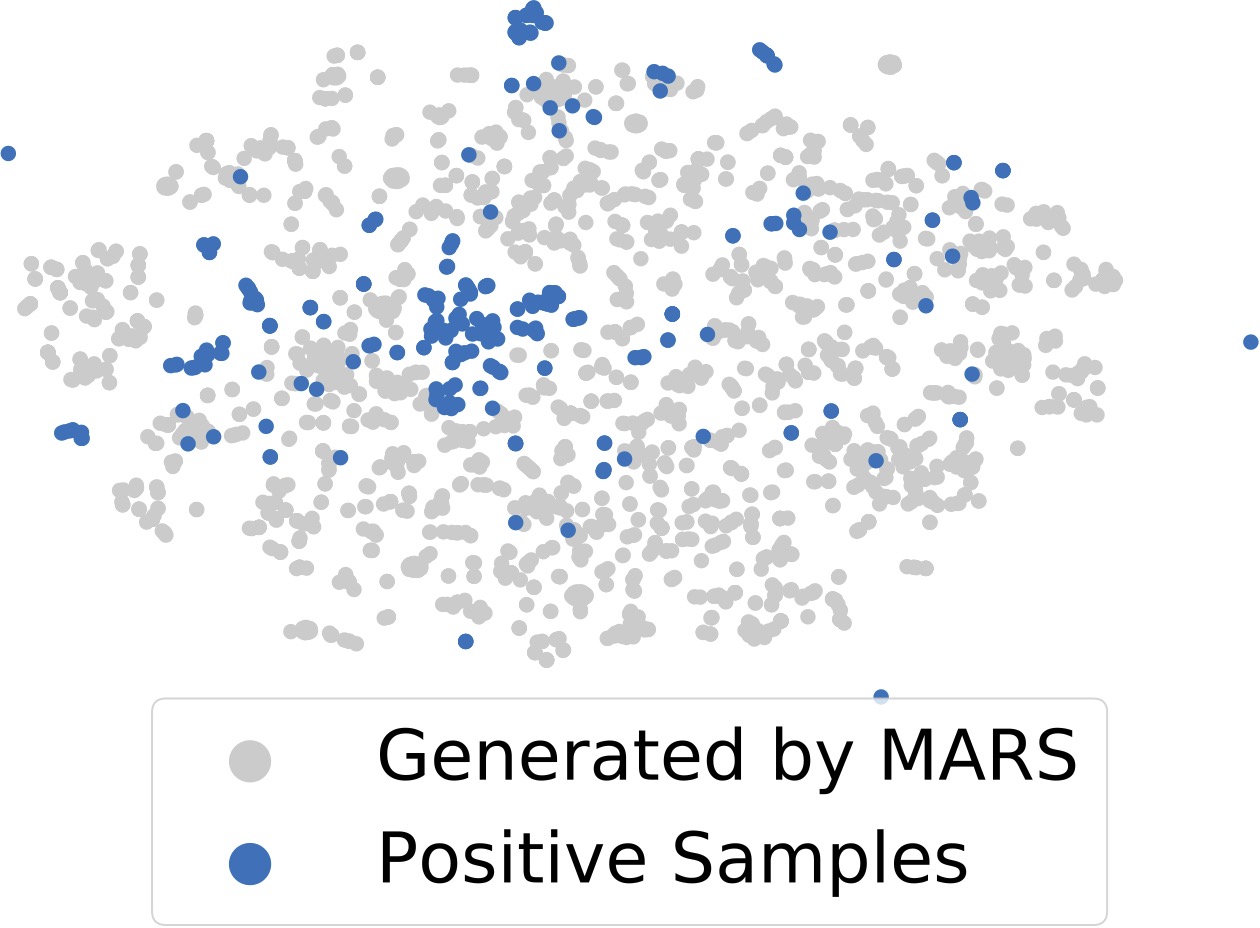
Shitong Luo*, Chence Shi*, Minkai Xu, Jian Tang 35th Conference on Neural Information Processing Systems (NeurIPS 2021) Molecular 3D geometry generation; long-range interactions; SE(3)-equivariance. [PDF] [Code] |
|
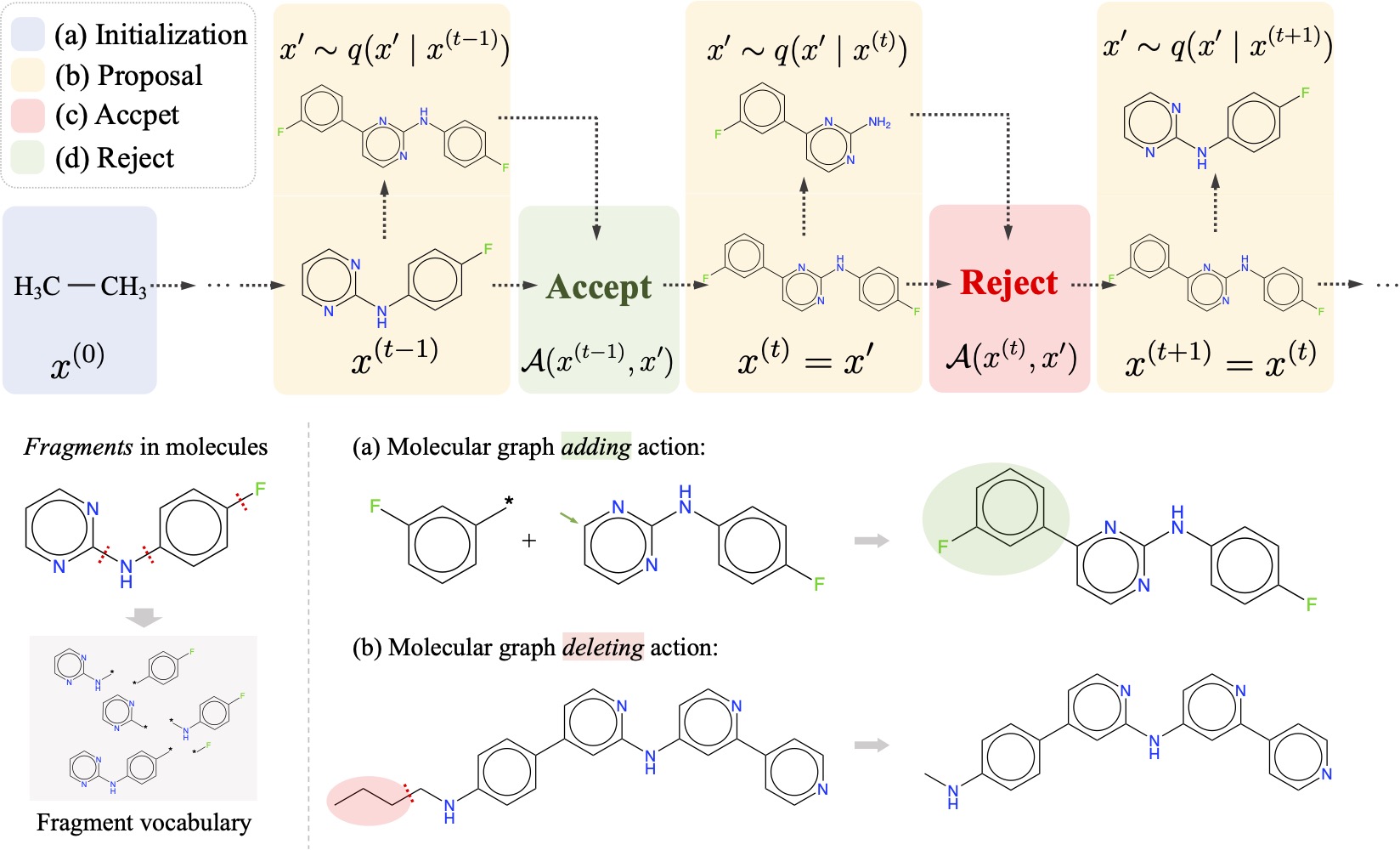
Yutong Xie, Chence Shi, Hao Zhou, Yuwei Yang, Weinan Zhang, Yong Yu, Lei Li 9th International Conference on Learning Representations (ICLR 2021) Multi-objective molecular graph generation via MCMC. Spotlight Presentation [PDF] [Code] |

|
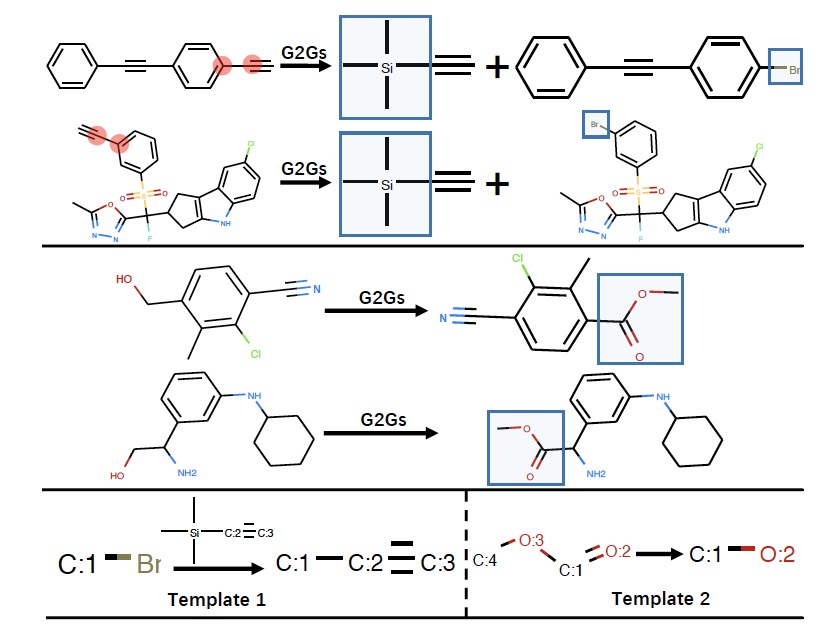
Chence Shi, Minkai Xu, Hongyu Guo, Ming Zhang, Jian Tang 37th International Conference on Machine Learning (ICML 2020) Retrosynthesis via graph translation; inspired by the disconnection approach in Organic Synthesis. [PDF] [Code] [Video Recording] [Slides] |
|
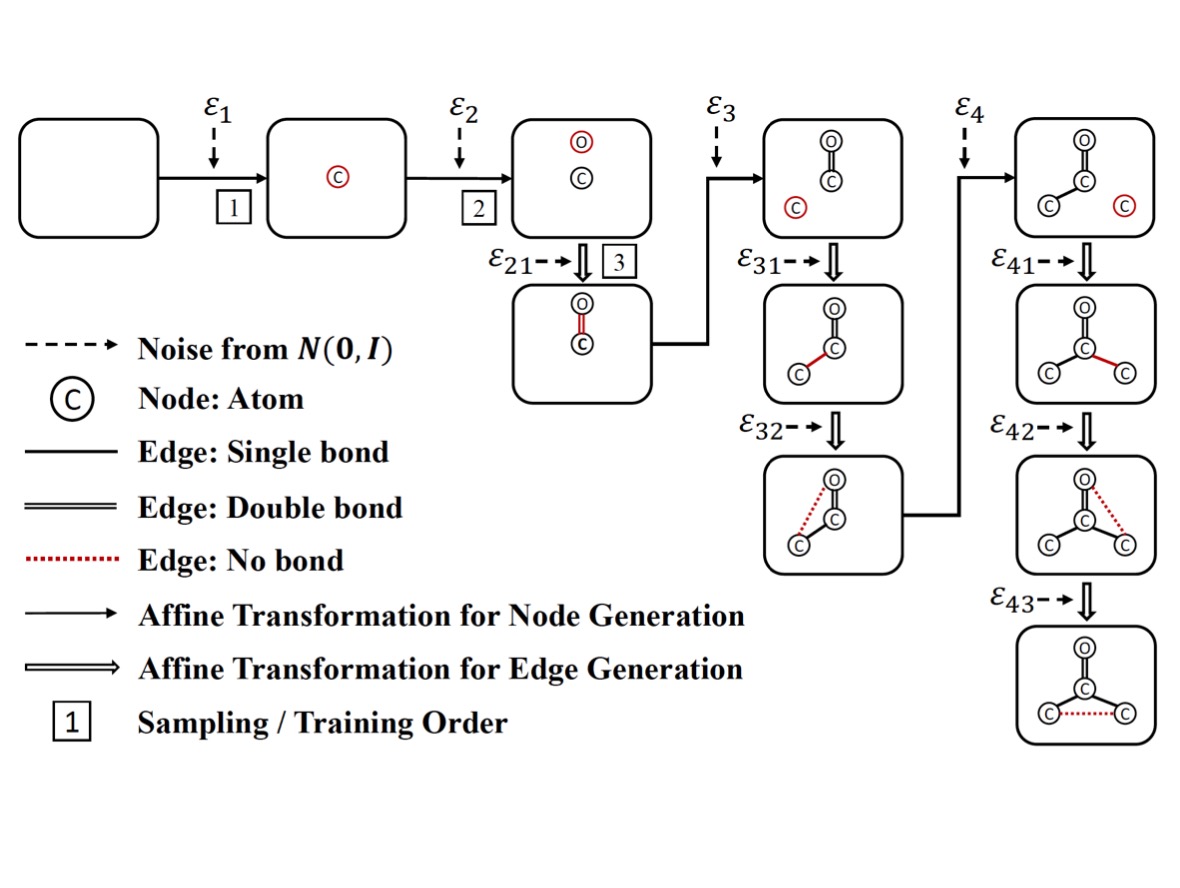
Chence Shi*, Minkai Xu*, Zhaocheng Zhu, Weinan Zhang, Ming Zhang, Jian Tang 8th International Conference on Learning Representations (ICLR 2020) Molecular graph generation and optimization; flow-based generative models. [PDF] [Code] [Video Recording] [Slides] |
|
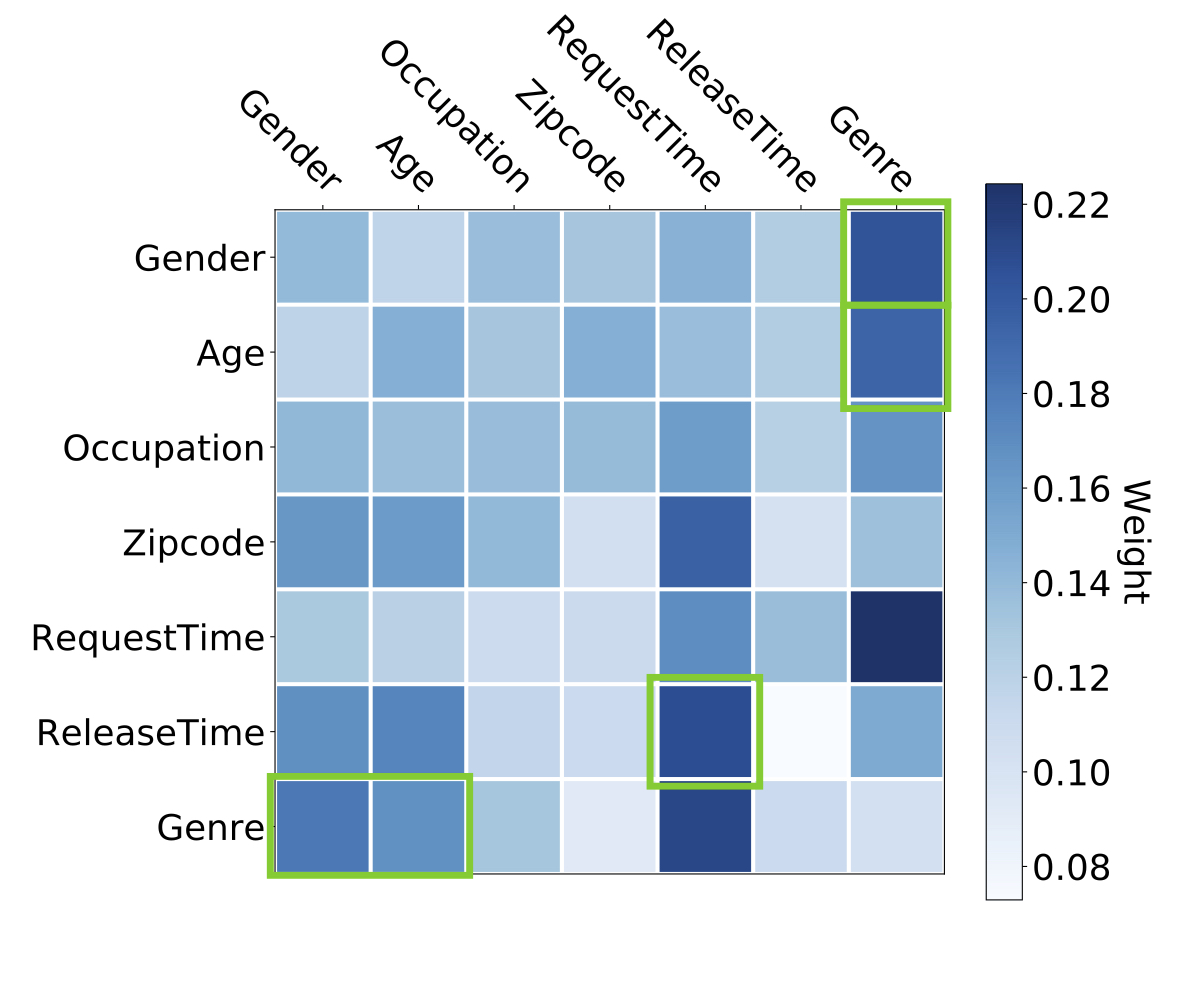
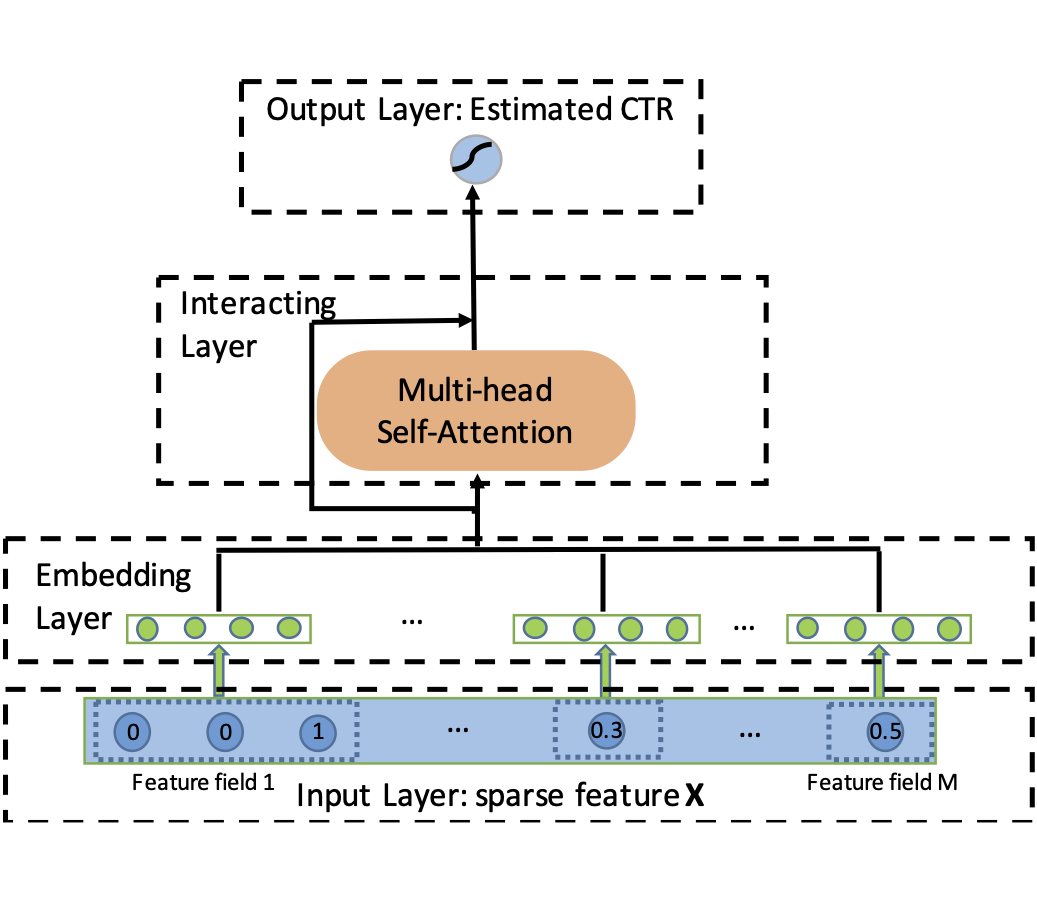
Weiping Song, Chence Shi, Zhiping Xiao, Zhijian Duan, Yewen Xu, Ming Zhang, Jian Tang 28th International Conference on Information and Knowledge Management (CIKM 2019) Click-through Rate Prediction with transformers. A well-known method in the industry [PDF] [Code] [Slides] |
|
(* equal contribution) |
|
• TorchDrug: A powerful and flexible machine learning platform for drug discovery.
• Recommender Systems: Code base on different recommendation topics, a comprehensive reading list and a set of bechmark data sets.
|
|
Program Committee member / Reviewer: ICML'21-23, NeurIPS'21-23, ICLR'22-23 |
|
Updated at Dec. 2024
|